SNPannotator
Adding physical and genetic position data to Affymetrix genotype files
When genotyping results are exported from the Affymetrix Genotyping Console version 2, the individual SNPs are no longer paired with their respective physical or genetic positions. Since these map positions are needed by the AutoSNPa and IBDfinder programs, such genotype files must first be annotated using SNPAnnotator.
The SNPannotator program first reads an annotation reference file, which can be downloaded (free of charge) from the Affymetrix web site. When the program has extracted the relevant positional data, it then reads the genotype text files in a folder and creates the annotated versions with *.xls file extensions.
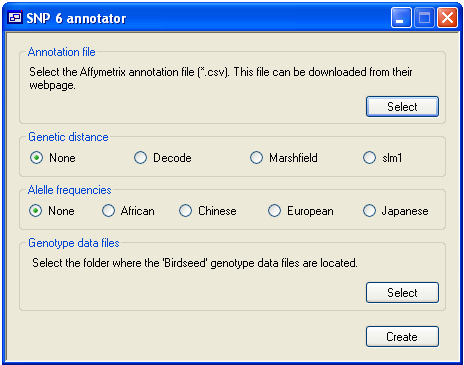 Each *.xls file contains the original genotype data linked to a SNP RS name, chromosome, and
physical position, as well as an optional genetic map position (Marshfield, Decode or SLM1). These files can then be used by any software package
that requires the data in the original Affymetrix file format.
Each *.xls file contains the original genotype data linked to a SNP RS name, chromosome, and
physical position, as well as an optional genetic map position (Marshfield, Decode or SLM1). These files can then be used by any software package
that requires the data in the original Affymetrix file format.
Download here (This program was updated on 28-Aug-2009, to read both the new and old formats of the annotation *.csv files, and now can also export allele frequency data.)
Note: this program requires the .NET framework version 2.0 to be installed (available through Windows Update).